- (2023) Eric Medwedeff and Eric Mjolsness, “Approximate Simulation of Cortical Microtubule Models using Dynamical Graph Grammars”, Physical Biology, v 20 no. 5, 7 July 2023.
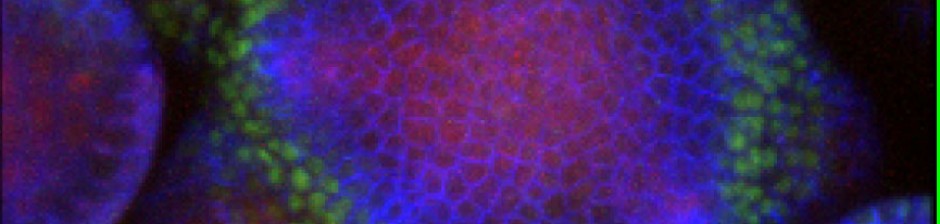
- (2023) Cory Scott, Eric Mjolsness, Diane Oyen, Chie Kodera, Magalie Uyttewaal, and David Bouchez, “Graph Metric Learning Quantifies Morphological Differences between Two Phenotypes of Shoot Apical Meristem Cells in Arabidopsis”. In Silico Plants, volume 5, issue 1, 30 January 2023, diad001. https://doi.org/10.1093/insilicoplants/diad001.
- (2022) Eric Mjolsness, “Explicit Calculation of Structural Commutation Relations for Stochastic and Dynamical Graph Grammar Rule Operators in Biological Morphodynamics”. Frontiers in Systems Biology, 09 September 2022. https://doi.org/10.3389/fsysb.2022.898858 . Also a book chapter in https://www.frontiersin.org/research-topics/30021/pattern-formation-in-biology (—>epub).
- (2021) C.B. Scott and Eric Mjolsness, “Graph diffusion distance: Properties and efficient computation”. PLoS One, https://doi.org/10.1371/journal.pone.0249624 April 27, 2021.
- (2019) C. B. Scott and Eric Mjolsness. Multilevel artificial Neural Network Training for Spatially Correlated Learning . SIAM J. Sci. Comput., vol 41 no. 5, pp. S297-S320, October 2019. https://doi.org/10.1137/18M1191506 .
- (2019) Mark Alber, Christophe Godin, Philip Maini, Roeland Merks, and Eric Mjolsness. Introduction. Editorial introduction to the Special Issue on Modeling Tissue Growth and Shape, Bulletin of Mathematical Biology, vol. 81 no. 8, August 2019. https://doi.org/10.1007/s11538-019-00649-2
- (2019) Oliver K. Ernst, Tom Bartol, Terrence Sejnowski, Eric Mjolsness. Learning Moment Closure Approximations in Reaction-Diffusion Systems with Spatial Dynamic Boltzmann Distributions. Physical Review E, v.99, 063315, 26 June 2019. DOI: 10.1103/PhysRevE.99.063315 .
- (2019) Eric Mjolsness. Prospects for Declarative Mathematical Modeling of Complex Biological Systems. Bulletin of Mathematical Biology, Volume 81, Issue 8, pp 3385–3420, August 2019. https://doi.org/10.1007/s11538-019-00628-7. Also preprint http://arxiv.org/abs/1804.11044 in a different format.
- (2019) Olivier Hamant, Daisuke Inoue, David Bouchez, Jacques Dumais, and Eric Mjolsness. Are microtubules tension sensors? Nature Communications, v10 article no. 2360, 29 May 2019.
- (2018) Oliver K. Ernst, Tom Bartol, Terrence Sejnowski, and Eric Mjolsness.Learning Dynamic Boltzmann Distributions as Reduced Models of Spatial Chemical Kinetics. Journal of Chemical Physics 149, 034107, July 2018. Also arXiv 1803.01063, March 2018.
- (2015) Bruce E. Shapiro and Eric Mjolsness. Pycellerator: An arrow-based reaction-like modelling language for biological simulations, Bioinformatics, Oct 26. 2015. DOI: 10.1093/bioinformatics/btv596.
- (2015) Todd Johnson, Thomas Bartol, Terrence Sejnowski, and Eric Mjolsness. Model Reduction for Stochastic CaMKII Reaction Kinetics in Synapses by Graph-Constrained Correlation Dynamics. Physical Biology 12:4, July 2015.
- (2015) Bruce E. Shapiro, Cory Tobin, Eric Mjolsness, and Elliot M. Meyerowitz. Analysis of Cell Divisions Patterns in the Arabidopsis Shoot Apical Meristem. Proceedings of the National Academy of Sciences 112:15 pp 4815–4820, 2015.
- (2013) “Using Cellzilla for Plant Growth Simulations at the Cellular Level”, Bruce E. Shapiro, Elliot Meyerowitz, Eric Mjolsness. Frontiers in Plant Biophysics and Modeling, 4:00408, 2013.
- (2013) “Model of Structuring the Stem Cell Niche in Shoot Apical Meristem of Arabidopsis thaliana”, S.V. Nikolaev, U.S. Zubairova, A.V. Penenko, E.D. Mjolsness, B.E. Shapiro, N.A. Kolchanov., Doklady Akademii Nauk, 2013, Vol. 452, No. 3, pp. 336–338, September 2013. Original in Russian. English version is Doklady Biological Sciences, 2013, Vol. 452, pp. 316–319. (Pleiades Publishing)
- (2013) Eric Mjolsness. Time-ordered product expansions for computational stochastic systems biology. Physical Biology 10 035009, 2013. [Journal paper, open access.]
- (2013) Eric Mjolsness and Upendra Prasad. Mathematics of small stochastic reaction networks: A boundary layer theory for eigenstate analysis, Journal of Chemical Physics 138, 104111 March 2013. [Journal Paper]
- (2012) David Orendorff and Eric Mjolsness. A hierarchical exact accelerated stochastic simulation algorithm, Journal of Chemical Physics 137, 214104, December 2012. DOI: 10.1063/1.4766353 ; arXiv:1212.4080. [Journal Paper | Technical Report]
- (2012) Eric Mjolsness, Compositional stochastic modeling and probabilistic programming. Workshop on Probabilistic Programming, Neural Information Processing Systems Conference Workshops, extended abstract, December 2012. [Mjolsness_1212.0582] Also available as [arXiv:1212.0582]
- (2012) Mironova VV, Novoselova ES, Doroshkov AV, Kazantsev FV, Omelyanchuk NA, Kochetov AV, Kolchanov NA, Mjolsness E. , Likhoshvai VA. Combined in silico/in vivo analysis of mechanisms providing for root apical meristem self-organization and maintenance , Annals of Botany 110:2 pp 349-360, July 2012; doi: 10.1093/aob/mcs069.
- (2012) Eric Mjolsness and Alexandre Cunha. “Topological object types for morphodynamic modeling languages”, PMA 2012 : IEEE Fourth International Symposium on Plant Growth Modeling, Visualization and Applications. Shanghai China, October 2012. IEEE Press. [Technical Report]
- (2011) Jonathan Young, James Locke, Alphan Altinok, Nitzan Rosenfeld, Tigran Bacarian, Peter Swain, Eric Mjolsness, and Michael Elowitz. Measuring single-cell gene expression dynamics in bacteria using fluorescence time-lapse microscopy, Nature Protocols 7:1, pp. 80-88, 2012. Published online December 15, 2011. [Paper]
- (2010) Mironova, V.V., Omelyanchuk, N.A., Yosiphon, G. Fadeev, S.I., Kolchanov, N.A., Mjolsness, E. and Likhoshvai, V.A. A plausible mechanism for auxin patterning along the developing root , BMC Systems Biology 4:98. [Published PDF ]
- (2010) Mjolsness, E. Towards Measurable Types for Dynamical Process Modeling Languages , To appear in Electronic Notes in Theoretical Computer Science (ENTCS), Elsevier. [ Author’s technical report version, PDF ]
- Wang, Y., Christley, S., Mjolsness, E., and Xie, X. Parameter inference for discretely observed stochastic kinetic models using stochastic gradient descent, BMC Systems Biology 4:99 . [ Published PDF ]
- (2010) Najdi, T.S., Hatfield, G. W. and Mjolsness, E. D. A `Random Steady State` Model for the Pyruvate Dehydrogenase and Alpha-Ketogluterate Dehydrogenase Enzyme Complexes , Physical Biology, 7 (2010) 016016 [ Author’s version, PDF | View published paper at publisher`s web site ]
- (2009) Golubyatnikov, V.P., Mjolsness, E., Gaidov, Yu.A. Topological index of the p53-Mdm2 circuit , The Herald of Vavilov Society for geneticists and breeding scientists (Informatzionnyi Vestnik Vavilovskogo obshchestva genetikov i selektzionerov) v. 13, N 1, 2009, pp 160 – 162. In English, 2009. [ Published PDF ]
- (2009) Mjolsness E, Orendorff D, Chatelain P, Koumoutsakos P. An Exact Accelerated Stochastic Simulation Algorithm, Journal of Chemical Physics 130:144110, 2009. Technical Report UCI-ICS 08-09 [Journal paper | View paper at publisher’s web site |Technical report ]
- (2008) Yosiphon G, Mjolsness E. Towards the Inference of Stochastic Biochemical Network and Parameterized Grammar Models, In N. Lawrence , M. Girolami, M. Rattray, and G. Sanguinetti eds., Learning and Inference in Computational Systems Biology, MIT Press, 2010. Also Technical Report UCI-ICS 08-07 [ Authors’ version, PDF ]
- (2007) Nikolaev SV, Penenko AV, Lavreha VV, Mjolsness ED, Kolchanov NA A Model Study of the Role of Proteins CLV1, CLV2, CLV3, and WUS in Regulation of the Structure of the Shoot Apical Meristem, Originally published in Russian, Ontogenez 2007, 38(6):457-462. [ Download PDF preprint ]
- (2007) Likhoshvai VA, Omel’yanchuk NA, Mironova VV, Fadeev SI, Mjolsness ED and Kolchanov NA Mathematical Model of Auxin Distribution in the Plant Root Russian Journal of Developmental Biology, 38(6):374-384; Original Russian Verison published in Ontogenez 38(6):446-456(2007). [ Download PDF ]
- (2007) Towards a Calculus of Biomolecular Complexes at Equilibrium . Eric Mjolsness, Briefings in Bioinformatics, 8(4):226-33 July 2007. [ Authors’ Version (Technical Report)| View Paper at Publisher’s Web Site ]
- (2007) On Cooperative Quasi-Equilibrium Models of Transcriptional Regulation. Eric Mjolsness, Journal of Bioinformatics and Computational Biology, vol 5 no 2(b) pp 467-490, 2007. Also Technical Report UCI-ICS 06-13, 2006. [ Technical Report | View Paper at Publisher’s Web Site ]
- (2006) Stochastic Process Semantics for Dynamical Grammars. Eric Mjolsness and Guy Yosiphon, Annals of Mathematics and Artificial Intelligence, 47(3-4) August 2006. [Authors’ Version (Technical Report) | View Paper at Publisher’s Web Site ]
- (2006) Connectivity in the Yeast Cell Cycle Transcription Network: Inferences from Neural Networks. Christopher Hart, Eric Mjolsness, Barbara Wold, PLOS Computational Biology vol. 2 no. 12, December 2006. [ Published paper, in PDF Site | Authors’ version]
- (2006) The Growth and Development of Some Recent Plant Models: A Viewpoint. Eric Mjolsness, Journal of Plant Growth Regulation, Journal of Plant Growth Regulation 25(4), 270-277, December 2006. [Authors’ version | View Paper at Publisher’s Web Site ]
- (2006) Static and dynamic models of biological networks. Ashish Bhan and Eric Mjolsness, Complexity , 11(6), 2006. [ View Paper at Publisher’s Web Site | Authors’ version ]
- (2006) Analysis of a one-dimensional model for the regulation of the size of the renewable zone in biological tissue. S.I. Fadeev, S.V. Nikolaev, V.V. Kogay, E. Mjolsness , N.A. Kolchanov, Computational Technologies, 11(2), 2006. [ | Authors’ version (In Russian) | Authors’ version (partial English translation) ]
- (2006) Application of a Generalized MWC Model for the Mathematical Simulation of Metabolic Pathways Regulated by Allosteric Enzymes. Tarek S. Najdi, Chin-Ran Yang, Bruce E. Shapiro, G. Wesley Hatfield, and Eric D. Mjolsness, Journal of Bioinformatics and Computational Biology, 4:335-355, 2006. [ View Paper at Publisher’s Web Site |Authors’ version ]
- (2006) An auxin-driven polarized transport model for phyllotaxis. Henrik Jönsson, Marcus G. Heisler, Bruce E. Shapiro, Elliot M. Meyerowitz, and Eric Mjolsness,Proceedings of the National Academy of Sciences, 13 January 2006. [ View Paper at Publisher’s Web Site | Authors’ version |PNAS Commentary | Supplementary Information | Cellerator Notebook ].
- (2005) The Generalized Monod, Wyman, Changeux model for mathematical modeling of metabolic enzymes with allosteric regulation. Tarek S. Najdi, Chin-Rang Yang, Bruce E. Shapiro, G. Wesley Hatfield and Eric D. Mjolsness, IEEE Computational Systems Bioinformatics Conference, Stanford University, August 8-12 2005. [ View Paper at Publisher’s Web Site ].
- (2005) Sigmoid: Towards an Intelligent, Scalable, Software Infrastructure for Pathway Bioinformatics and Systems Biology . Jianlin Cheng, Lucas Scharenbroich, Pierre Baldi, Eric Mjolsness, IEEE Intelligent Systems , May/June 2005. [ Journal article ]
- (2005) A Mathematical Model for the Branched Chain Amino Acid Biosynthetic Pathways of Escherichia coli K12 . Chin-Ran Yang, Bruce E. Shapiro, She-pin Hung, Eric D. Mjolsness, and G. Wesley Hatfield, Journal of Biological Chemistry, 2005 Mar 25;280(12):11224-32 . [ Authors’ version | View Paper at Publisher’s Web Site ].
- (2005) An enzyme mechanism language for the mathematical modeling of metabolic pathways . Yang, C-R., Shapiro, B.E., Mjolsness, E.D., and Hatfield, G.W..Bioinformatics, vol. 21 no. 6, pages 774-780, March 2005. [ Authors’ version | View Paper at Publisher’s Web Site ].
- (2005) Modeling the organization of the WUSCHEL expression domain in the shoot apical meristem. Jönsson H, Heisler M, Reddy GV, Agrawal V, Gor V, Shapiro BE, Mjolsness E, and Meyerowitz E.M. Bioinformatics 21(S1): i232-i240.[ View Abstract or Paper at Publisher’s Web Site. | Supplementary Material | PDF ]
- (2005) A mathematical and computational framework for quantitative comparison and integration of large scale gene expression data. Hart CE, Sharenbroich L, Bornstein BJ, Trout D. King B., Mjolsness E., and Wold BJ. Nucleic Acids Research, May 10;33(8):2580-94, 2005. [ Abstract | Paper ]
- (2005) Static and Dynamic Models of Biological Networks. Ashish Bhan and Eric Mjolsness, UCI ICS TR# 05-15. December 2005. [ Technical Report ]
- (2005) Variable-structure systems from graphs and grammars. Eric Mjolsness, UCI ICS TR# 05-09. July 2005. [Technical Report] [old link to Technical Report ]
- (2004-05) A Software architecture for developmental modeling in plants: The Computable Plant project. Gor V, Shapiro BE, Jönsson H, Heisler M, Reddy GV,Meyerowitz EM, Mjolsness E, Presented at the Fourth International Conference on Bioinformatics of Genome Regulation and Structure (BGRS-2004) , Novosibirsk, Russia, July 2004. In: Bioinformatics of Genome Regulation and Structure II, (Eds. N.Kolchanov and R. Hofestaedt) Springer Science+Business Media, Inc. 2005, pp. 345-354. [Abstract | Preprint ]
- (2004) Labeled graph notations for graphical models: Extended Report. Eric Mjolsness, UCI ICS TR# 04-03. March 2004. [ Technical Report ]
- (2004) Modeling plant development with gene regulation networks including signaling and cell division. Jönsson, H., Shapiro, B.E., Meyerowitz, E.M., and Mjolsness, E. , inBioinformatics of Genome Regulation and Structure, eds R. Hofestaedt and N. Kolchanov, Kluwer Publications, pp 311-318, 2004. [ Abstract ]
- (2003) Signaling in multicellular models of plant development. Jönsson, H., Shapiro, B.E., Meyerowitz, E.M. and Mjolsness, E. In S. Kumar and P.J. Bentley (Eds.) On Growth, Form, and Computers, Academic Press, London, UK, pages 156-161. [ Author’s copy | Abstract |Publisher’s Web Site ]
- (2003) Cellerator: extending a computer algebra system to include biochemical arrows for signal transduction simulations. Shapiro, B.E., Levchenko, A., Meyerowitz, E.M., Wold, B.J., and Mjolsness, E.D. Bioinformatics, 19(5):677-678. [ View Abstract or Paper at Publisher’s Web Site | PDF ]
- (2003) Gene Expression Clustering with Functional Mixture Models. Padhraic Smyth, Darya Chudova, Christopher Hart, Eric Mjolsness, Advances in Neural Information Processing Systems, 2003. NIPS2003_AP15 [ Preprint ]
- (2003) The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. M. Hucka, A. Finney, H. M. Sauro, H. Bolouri, J. C. Doyle, H. Kitano , A. P. Arkin, B. J. Bornstein, D. Bray, A. Cornish-Bowden, A. A. Cuellar, S. Dronov, E. D. Gilles, M. Ginkel, V. Gor, I. I. Goryanin, W. J. Hedley, T. C. Hodgman, J.-H. Hofmey, P. J. Hunter, N. S. Juty, J. L. Kasberger, A. Kremling, U. Kummer, N. Le Nov` ere, L. M. Loew, D. Lucio , P. Mendes, E. Minch, E. D. Mjolsness, Y. Nakayama, M. R. Nelson, P. F. Nielsen, T. Sakurada, J. C. Schaff , B. E. Shapiro, T. S. Shimizu, H. D. Spence, J. Stelling. Bioinformatics Vol 19, no 4, 2003 pp 524-531. [ View Abstract or Paper at Publisher’s Web Site ]
- (2003) Gene Network Models and Neural Development. George Marnellos and Eric D. Mjolsness. In Modeling Neural Development, edited by Arjen van Ooyen. The MIT Press, Cambridge, Massachusetts, 2003. [ Preprint ]
- (2002) Modeling plant development with gene regulation networks including signaling and cell division. Mjolsness E, Jönsson H, Shapiro BE, Meyerowitz EM. Paper presented at the Third International Conference on Bioinformatics of Genome Regulation and Structure (BGRS 2002), Novosibirsk, Russia, July 14-20, 2002. [ Abstract | Paper |Conference Proceedings ].
- (2002) Automatic model generation for signal transduction with applications to map kinase pathways. Shapiro BE, Levchenko A, Mjolsness E. In: Foundations of Systems Biology, H. Kitano, ed, MIT Press, pages 145-162) ISBN 0-262-11266-3 [ Publisher’s web site | Preprint ]
- (2001) Developmental simulations with Cellerator. Shapiro BE, Mjolsness ED. Paper presented at the Second International Conference on Systems Biology, Pasadena, CA, Novemeber 2001. [ Preprint ]
- (2001) Gene regulation networks for modeling Drosophila development. Eric Mjolsness, in Computational Methods in Molecular Biology, eds. J. M. Bower and H. Bolouri, MIT Press 2001. [ Preprint ]
- (2001) Modeling the activity of single genes. Michael Gibson and Eric Mjolsness, inComputational Methods in Molecular Biology, eds. J. M. Bower and H. Bolouri, MIT Press 2001. [ Preprint ]
- (2000) Delta-Notch lateral inhibitory patterning in the emergence of ciliated cells in Xenopus: experimental observations and a gene network model.. Marnellos G, Deblandre GA, Mjolsness E, Kintner C. Pacific Symposium on Biocomputing pp. 329-40 2000. [ Abstract | Abstract | Paper ]
- (1999) From Coexpression to Coregulation: An Approach to Inferring Transcriptional Regulation among Gene Classes from Large-Scale Expression Data. E. Mjolsness, T. Mann, R. Castano, and B. Wold. Advances in Neural Information Processing Systems 1999. [ Paper | Paper]
- (1998) A Gene Network Model of Resource Allocation to Growth and Reproduction. G. Marnellos and E. Mjolsness, Artificial Life VI, eds. C. Adami, R. K. Belew, H. Kitano, and C. E. Taylor, MIT Press, 1998. [ Preprint | Preprint]
- (1998) Probing the Dynamics of Cell Differentiation in a Model of Drosophila Neurogenesis. G. Marnellos and E. Mjolsness, Artificial Life VI, eds. C. Adami, R. K. Belew, H. Kitano, and C. E. Taylor, MIT Press, 1998. [ Preprint ]
- (1998) A Gene Network Approach to Modeling Early Neurogenesis in Drosophila. G. Marnellos and E. Mjolsness, in Pacific Symposium on Biocomputing, eds. R. B. Altman, A. K. Dunker, L. Hunter and T. Klein, World Scientific, 1998. [ Preprint ]
- (1998) A Lagrangian formulation of neural networks I: Theory and analog dynamics. Eric Mjolsness and Willard L. Miranker,
Neural, Parallel and Scientific Computations vol 6 no. 3, pp. 297-336, 1998.
[Preprint: I] [ Scanned journal PDF – 13MB ]
- (1998) A Lagrangian Formulation of Neural Networks II: Clocked Objective Functions and Applications, Willard L. Miranker and Eric Mjolsness, Neural, Parallel and Scientific Computations, vol 6 no. 3, pp. 337-372, 1998. [Preprint of Parts I and II together]
- (1997) Symbolic Neural Networks Derived from Stochastic Grammar Domain Models. Eric Mjolsness, in Connectionist Symbolic Integration , eds. R. Sun and F. Alexandre, Lawrence Erlbaum Associates, 1997. [ Preprint ]
- (1995) Model for cooperative control of positional information in Drosophila by bcd and maternal hb. John Reinitz, Eric Mjolsness and David H. Sharp, Journal of Experimental Zoology 271:47-56, 1995. [ Abstract | Paper | Preprint]
- (1995) Modeling the Connection between Development and Evolution: Preliminary Report. Eric Mjolsness, Charles D. Garrett, John Reinitz, and David H. Sharp, in Evolution and Biocomputation: Computational Models of Evolution eds. Wolfgang Banzhaf and Frank H. Eeckman, Lecture Notes in Computer Science, Springer, Berlin, 1995. [ Paper ]
- (1993) Multiscale Models of Developmental Processes. David H. Sharp, John Reinitz, and Eric Mjolsness, in Open Systems and Information Dynamics, vol 2 no 1, pp. 1-10, Nicholas Copernicus University Press, Torun, Poland, 1993. [Paper]
- (1993) Animation of plant development Przemyslaw Prusinkiewicz, Mark S. Hammel, and Eric Mjolsness, SIGGRAPH ’93 Conference Proceedings, ACM 1993. [ Preprint ]
- (1992) A Connectionist Model of the Drosophila Blastoderm. J. Reinitz, E. Mjolsness, and D. H. Sharp, in The Principles of Organization in Organisms , eds. Jay E. Mittenthal and Arthur B. Baskin, Santa Fe Institute Studies in the Sciences of Complexity, Addison-Wesley 1992. [Preprint, technical report LA-UR–90-3923.]
- (1992) Visual grammars and their neural nets Eric Mjolsness, Advances in Neural Information Processing Systems 4, editors Moody, Hanson and Lippmann, pp. 428-435, Morgan-Kaufman 1992. [ Paper ]
- (1991) A Lagrangian Approach to Fixed Points. Eric Mjolsness and Willard L. Miranker,Advances in Neural Information Processing Systems 3, editors Lippmann, Moody and Touretsky, Morgan-Kaufman 1991. [ Paper ]
- (1991) Multiscale optimization in neural networks, Eric Mjolsness, Charles Garrett, and Willard Miranker, IEEE Transactions on Neural Networks , vol 2 no 2, March 1991. [Paper ]
- (1991) A connectionist model of development. Mjolsness E, Sharp DH, Reinitz J. Journal of Theoretical Biology 152: 429-453. [ Abstract | Paper ]
- (1990) A connectionist Model of the Drosophila Blastoderm. John Reinitz, Eric Mjolsness, and David H. Sharp, Report LA-UR–90-3923, Los Alamos National Laboratory, November 1990. [ Technical Report, Scanned PDF ]
- (1990) Bayesian inference on visual grammars by neural nets that optimize. Eric Mjolsness, Report YALEU/DCS/TR- 854, May 1990. [ Technical Report ]
|